
|
个人简介Personal Profile
个人简介
黄康,教授,博士生导师。
研究方向:行为生态学、分子生态学、进化生态学、群体遗传学。
主要围绕动物学、生态学相关领域开展研究工作,以秦岭川金丝猴为模型,研究物种隔离分化的成因机制、动物亲选择行为的影响因素、复杂社会体系的形成机制等问题;将数学、统计学和计算机同生态学问题相结合,开发生态学模型和分析方法。
教育及工作经历
2023.1 至今 西北大学 生命科学学院 教授
2017.5-2022.12 西北大学 生命科学学院 副教授
2015.3-2017.4 西北大学 生命科学学院 讲师
2018.9-2019.8 加拿大UBC大学 林学院 访问学者
2010.9-2015.1 西北大学 动物学 博士生
2009.9-2010.7 西北大学 动物学 硕士生
2005.9-2009.7 西北大学 生物科学 本科生
讲授课程
本科二年级《生物统计学》
本科三年级《生态学数据分析》
硕士一年级《生物统计学与数据分析》
人才计划
2020年陕西省青年科技新星
2017年西北大学青年学术英才支持计划
2017年陕西省高校首批“青年杰出人才支持计划”
2017年中国科协青年人才托举工程(国际动物学会推荐)
获奖情况
2021年优秀青年动物生态学工作者
2020年中国动物学会长隆奖-启航奖
2017年陕西省优秀博士学位论文
2017年陕西省第十三届自然科学优秀学术论文一等奖(排名3)
2014年陕西省首届研究生创新成果展一等奖
2013年陕西省第十二届自然科学优秀学术论文三等奖(排名2)
社会兼职
2022.12-2026.12,中国生态学学会动物生态专业委员会委员
2022.1-2026.1,中国动物学会进化理论专业委员会委员
2017.8-2026.7,中国动物学会灵长类学分会理事
2024.1-2027.12,陕西动物学会理事
科研项目
2026-2029,国家自然科学基金面上项目,川金丝猴的欺骗性警报及响应策略的研究,32570568,主持。
2022-2025,国家自然科学基金面上项目,相关样本的群体遗传学分析,32170515,主持。
2018-2021,国家自然科学基金面上项目,基于共显性遗传标记的同源多倍体进化生态学分析方法的研究,31770411,主持
2018-2019,陕西省自然科学基础研究计划面上项目,同源多倍体进化生态学分析方法的研究,2018JM3024,主持
2016-2018,国家自然科学基金青年项目,人为干扰和景观格局导致秦岭川金丝猴遗传分化机制的研究,31501872,主持
2015-2017,西北大学科研启动基金,小尺度下物种隔离因素的研究,主持
2018-2022,国家自然科学基金重点项目,川金丝猴全雄群社会系统的形成与维持机制的研究,31730104,第3参与人
2017-2020,国家自然科学基金面上项目,金丝猴重层社会中雌性在食物竞争中的合作策略研究,31672301,第2参与人
2015-2018,国家自然科学基金面上项目,秦岭川金丝猴社群分离聚合机制的研究,31470455,第5参与人
2013-2016,国家自然科学基金面上项目,秦岭川金丝猴营养需求和采食策略的研究,31270441,第3参与人
2012-2016,国家自然科学基金重点项目,川金丝猴社会体系稳定机制的研究,31130061,第10参与人
科研软件
1. VCFPOP: 用普通台式机完成100G级别二倍体、多倍体测序基因型文件的群体遗传学分析
2. POLYGENE:使用同源多倍体分子表型数据进行各种群体遗传学分析
3. PLOIDYINFER,从等位基因表型推断个体倍性
4. POLYRELATEDNESS:估计多倍体、不同倍性个体以及杂倍体的亲缘系数
5. ISOLATION:景观遗传学分析软件,计算最低成本路径、等效电阻和进行高阶偏Mantel检验
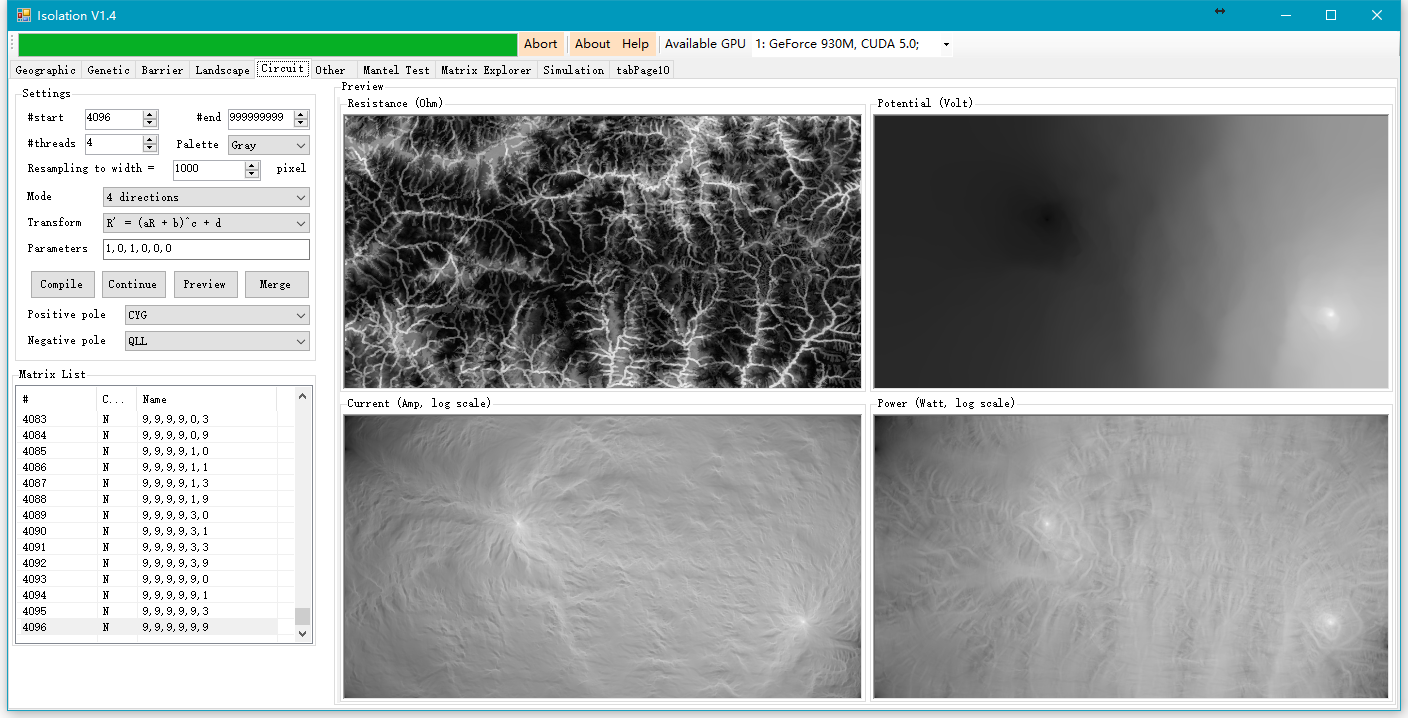
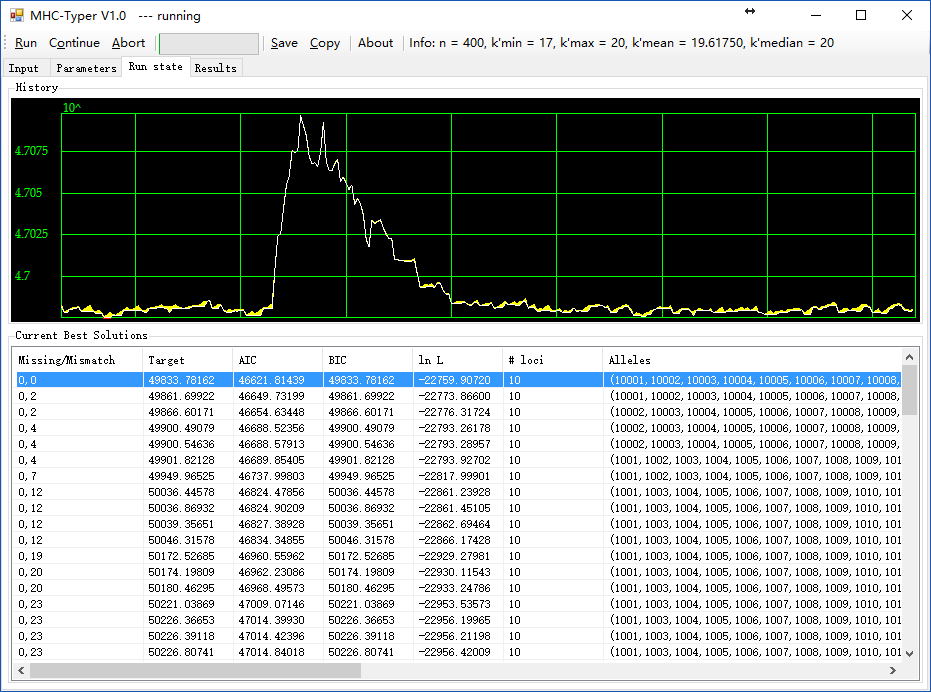
6. MHC-Typer:将多位点共同扩增的等位基因归入各自位点
软件下载:https://github.com/huangkang1987/
研究成果
Li WK#, Wang D#, Yang B, Shen YJ, Dunn DW, Hai X, Huang K*, Li BG (2026) Subgenome phasing reveals the asymmetric evolution and origin of the allotetraploid Achillea wilsoniana, Molecular Ecology, 35, e70237. IF=3.900.
Pan H, Guo GG, Wang J, He G, Guo ST, Zhang P, Hou R, Fang G, Li YL, Pan RL, Huang K*, Li BG* (2025) Amphibian biodiversity and distribution changes from the Paleozoic in China. Journal of Zoological Systematics and Evolutionary Research, 2025, 1176395. IF=2.600.
Pan H, Huang K, Wang J, Song CR, Guo F, Zhang X, Hu ZF, Zhang H, Ji KX, He SJ, Guo ST, Li DY, Tian WY, Zhao HT, Yue JB, Pan RL, Zhou ZH, He G*, Li BG* (2025) Evolutionary distribution changes of Sichuan golden monkeys (Rhinopithecus roxellana) in the Quaternary. Ecology and Evolution, 15, e72036. IF=2.300.
Wang D, Li WK, Yang B, Shen YJ, He J, Dunn DW, Huang K*, Li BG (2025) Epilobieae genomes and the evolution of Myrtales. BMC Plant Biology, 25, 950. IF=4.800.
Huang K, Li WK, Yang B, Wang D, He SJ, Shen YJ, Ao JC, Li YH, Cui YX, Kong YC, Li W, Li NL, Dunn DW, Li BG* (2025) vcfpop: performing population genetics analyses for autopolyploids and aneuploids based on next-generation sequencing datasets. Molecular Ecology Resources, 25, e13744. IF=8.678.
Pan H, Ji XP, Youlatos D, Chen Y, Zhang H, Guo GG, Wang J, Huang K, Hou R, He G, Guo ST, Zhang P, Li BG*, Pan RL* (2025) Morphometric study on the mandible of Colobine fossil (Mesopithecus pentelicus) found in East Asia, a comparison with extant taxa. American Journal of Primatology, 87, e23706. IF=3.014.
Dong SX, Zhang BY, Huang K, Ying MJ, Yan JB, Niu F, Hu HY, Dunn DW, Ren Y, Li BG*, Zhang P* (2024) Balancing selection shapes population differentiation of major histocompatibility complex genes in wild golden snub-nosed monkeys. Current Zoology, 70, 596-606. IF=2.200.
Li W, Dong SX, Niu F, Li NL, Su ZY, Wang CL, Huang K, Zhang H, Hou R, Wu T, Zhao HT, Wang Y, Wang XW, Pan RL*, Zhang P*, Li BG* (2024) Infanticide in golden snub-nosed monkeys with multilevel society. Current Zoology, 70, 273-275. IF=2.200.
Zhang P#*, Zhang B#, Dunn DW, Song XY, Huang K, Dong SX, Niu F, Ying MJ, Zhang YY, Shang YX, Pal RL, Li BG* (2023) Social and paternal female choice for male MHC genes in golden snub‐nosed monkeys (Rhinopithecus roxellana). Molecular Ecology, 32, 3239-3256. IF=6.622.
Wang TT#, Kong YC#, Zhang H, Li YH, Hou R, Dunn DW, Hou XD, Huang K*, Li BG (2023) Do golden snub-nosed monkeys use deceptive alarm calls during competition for food? iScience, 26, 106098. IF=6.107.
Li BG*, Zhang H, Huang K, He G, Guo ST, Hou R, Zhang P, Wang HT, Pan H, Fu HG, Wu XY, Jiang KX, Pan RL* (2022) Regional fauna-flora biodiversity and conservation strategy in China. iScience, 25, 104897. IF=6.107.
Zhang H, Lu JQ, Tang SY, Huang ZP, Cui LW, Lan DY, Wang HT, Hou R, Xiao W, Guo ST, He G, Huang K, Zhang P, Pan H, Oxnard CR, Pan RL*, Li BG* (2022) Southwest China, the last refuge of continental primates in East Asia. Biological Conservation, 273, 109681. IF=5.991.
Huang K, Dunn DW, Li WK, Wang D, Li BG* (2022) Linkage disequilibrium under polysomic inheritance. Heredity, 128, 11-20. IF=3.821.
Yang X, Berman CM, Hu HY, Hou R, Huang K, Wang XW, Zhao HT, Wang CL, Li BG*, Zhang P* (2022) Female preferences for male golden snub-nosed monkeys vary with male age and social context. Current Zoology, 68, 133-142. IF=2.624.
Huang K#, Zhang H#, Wang CL, Hou R, Zhang P, He G, Guo ST, Tang SY, Li BG*, Oxnard C, Pan RL* (2021) Use of historical and contemporary distribution of mammals in China to inform conservation. Conservation Biology, 35, 1787-1796. IF=6.560.
Huang K, Huber G, Ritland K, Dunn DW, Li BG* (2021) Performing parentage analysis for polysomic inheritances based on allelic phenotypes. G3: Genes, Genomes, Genetics, 11, jkaa064. IF=2.781
Huang K, Wang TT, Dunn DW, Zhang P, Sun HJ, Li BG* (2021) A generalized framework for AMOVA with multiple hierarchies and ploidies. Integrative Zoology, 16, 33-52. IF=2.514.
Hou R, Chapman CA, Rothman JM, Zhang H, Huang K, Guo ST*, Li BG*, Raubenheimer D (2021) The geometry of resource constraint: an empirical study of the golden snub-nosed monkey. Journal of Animal Ecology, 90, 751-765. IF=4.554.
Li YL#, Huang K#, Tang SY, Feng L, Yang J, Li ZH, Li BG* (2021) Genetic structure and evolutionary history of Rhinopithecus roxellana in Qinling Mountains, Central China. Frontiers in Genetics, 11, 611914. IF=3.258.
Huang K, Dunn DW, Ritland K, Li BG* (2020) POLYGENE: population genetics analyses for autopolyploids based on allelic phenotypes. Methods in Ecology and Evolution, 11, 448-456. IF=7.099.
Zhang BY, Hu HY, Song CM, Huang K, Dunn DW, Yang X, Wang XW, Zhao HT, Wang CL, Zhang P*, Li BG* (2020) MHC-based mate choice in wild golden snub-nosed monkeys. Frontiers in Genetics, 11, 609414. IF=3.258.
Li BG*, He G, Guo ST, Hou R, Huang K, Zhang P, Zhang H, Pan RL*, Chapman CA (2020) Macaques in China: Evolutionary dispersion and subsequent development. American Journal of Primatology, 82, e23142. IF=2.067.
Huang K#, Zhang P#, Dunn DW, Wang TC, Mi R, Li BG* (2019) Assigning alleles to different loci in amplifications of duplicated loci. Molecular Ecology Resources, 19, 1240-1253. IF=7.059.
Huang K, Dunn DW, Li ZH, Zhang P, Dai Y, Li BG* (2019) Inference of individual ploidy level using codominant markers. Molecular Ecology Resources, 19, 1218-1229. IF=7.059.
Huang K, Wang TC, Dunn DW, Zhang P, Cao XX, Liu RC, Li BG* (2019) Genotypic frequencies at equilibrium for polysomic inheritance under double-reduction. G3: Genes, Genomes, Genetics, 9, 1693-1706. IF=2.743.
Mi R, Wang TC, Dunn DW, Huang K*, Li BG (2019) Development of simple sequence repeat markers for Chamerion angustifolium (Onagraceae). Applications in Plant Sciences, 7, e1244. IF=1.187.
Huang K#, Mi R#, Dunn DW, Wang TC, Li BG* (2018) Performing parentage analysis in the presence of inbreeding and null alleles. Genetics, 210, 1467-1481. IF=4.075.
Zhang P#, Huang K#, Zhang BY, Dunn DW, Chen D, Li F, Qi XG, Guo ST, Li BG* (2018) High polymorphism in MHC-DRB genes in golden snub-nosed monkeys reveals balancing selection in small, isolated populations. BMC Evolutionary Biology, 18, 29. IF=3.027.
Ren Y#, Huang K#, Guo ST*, Pan RL, Dunn DW, Qi XG, Wang XW, Wang CL, Zhao HT, Yang B, Li FF, Li BG* (2018) Kinship promotes affiliative behaviors in a monkey. Current Zoology, 64, 441-447. IF=2.181.
Qi XG*, Huang K, Fang G, Grueter CC, Dunn DW, Li YL, Ji WH, Wang XY, Wang RT, Garber PA, Li BG* (2017) Male cooperation for breeding opportunities contributes to the evolution of multilevel societies. Proceedings of the Royal Society B: Biological Sciences, 284:20171480. IF=4.940.
Duan YM#, Huang K#, Qi XG*, Li BG* (2017) Characterization of 29 highly polymorphic microsatellite markers for golden snub-nosed monkey (Rhinopithecus roxellana). Conservation Genetics Resources, 9, 229-232. IF=0.446.
Huang K, Ritland K, Dunn DW, Qi XG, Guo ST, Li BG* (2016) Estimating relatedness in the presence of null alleles. Genetics, 202, 247-260. IF=5.963.
Huang K#, Guo ST#, Cushman SA, Dunn DW, Qi XG, Hou R, Zhang J, Li Q, Zhang Q, Shi Z, Zhang K, Li BG* (2016) Population structure of the golden snub-nosed monkey Rhinopithecus roxellana in the Qinling Mountains, central China. Integrative Zoology, 11, 350-360. IF=1.904.
Song XY#, Zhang P#, Huang K, Chen D, Guo ST, Qi XG, He G, Pan RL, Li BG* (2016) The influence of positive selection and trans-species evolution on DPB diversity in the golden snub-nosed monkeys (Rhinopithecus roxellana). Primates, 57, 489-499. IF=1.337.
Zhang P, Song X, Dunn DW, Huang K, Pan RL, Chen D, Guo ST, Qi XG, He G, Li BG* (2016) Diversity at two genetic loci associated with the major histocompatibility complex in the golden snub-nosed monkey (Rhinopithecus roxellana). Biochemical Systematics & Ecology, 68, 243-249. IF=0.988.
Yang B#, Zhang P#, Huang K, Garber PA, Li BG* (2016) Daytime birth and postbirth behavior of wild Rhinopithecus roxellana in the Qinling Mountains of China. Primates, 57, 155-160. IF=1.337.
Huang K, Ritland K, Guo ST, Dunn DW, Chen D, Ren Y, Qi XG, Zhang P, He G, Li BG* (2015) Estimating pairwise relatedness between individuals with different levels of ploidy. Molecular Ecology Resources, 15, 772-784. IF=5.626.
Huang K#, Guo ST#, Shattuck MR, Chen ST, Qi XG, Zhang P, Li BG* (2015) A maximum-likelihood estimation of pairwise relatedness for autopolyploids. Heredity, 114, 133-142. IF=3.804.
Guo ST#, Huang K#, Ji WH, Garber PA, Li BG* (2015) The role of kinship in the formation of a primate multilevel society. American Journal of Physical Anthropology, 156, 606-613. IF=2.514.
Huang K, Ritland K, Guo ST, Shattuck M, Li BG* (2014) A pairwise relatedness estimator for polyploids. Molecular Ecology Resources, 14, 734-744. IF=7.432.
Qi XG*, Garber PA, Ji WH, Huang ZP, Huang K, Zhang P, Guo ST, Wang XW, He G, Zhang P, Li BG* (2014) Satellite telemetry and social modeling offer new insights into the origin of primate multilevel societies. Nature Communications, 5, 5296. IF=10.742.
Wei W#, Qi XG#, Guo ST, Zhao DP, Zhang P, Huang K, Li BG* (2012) Market powers predict reciprocal grooming in golden snub-nosed monkeys (Rhinopithecus roxellana). PLoS ONE, 7, e36802. IF=4.092.
He G#, Huang K#, Guo ST*, Ji WH, Qi XG, Ren Y, Jin XL, Li BG* (2011) Evaluating the reliability of microsatellite genotyping from low-quality DNA templates with a polynomial distribution model. Chinese Science Bulletin, 56, 2523-2530. IF=1.087.

